The open-source tool Vertex AI AlphaFold Inference Pipeline has enabled biotech companies in streamlining protein-folding activities, accelerating their go to market timeline. It addresses key challenges in protein structure prediction by unleashing the power of parallel processing, optimizing compute resources, and scaling to meet high-throughput demands. Furthermore, it ensures reproducibility, lineage analysis, flexibility, adaptability, and seamless integration with upstream and downstream systems – all within Vertex AI as the one-stop platform. With this tool, researchers can unlock new possibilities, make groundbreaking discoveries faster than ever before, and drive end-to-end efficiency in their biotech drug discovery efforts.
However, even with Google Cloud’s efforts to make the AlphaFold algorithm more accessible to biotech firms, many bioscience organizations still struggle to integrate this technology seamlessly into their researchers’ workflows.
The biggest challenge is this: scientists who obsess over protein shapes aren’t usually coding ninjas or cloud wizards. Asking them to wrestle with complicated setups just to get a glimpse of a protein is like asking a chef to build their own oven before they can cook dinner. It’s not the best recipe for success (or tasty results).
Solution Overview
To reduce the friction, we are making our Vertex AI AlphaFold Inference Pipeline easier to use, including introducing a user-friendly AlphaFold Portal – think of it like protein modeling for beginners. We empower scientists, irrespective of their prior experience with cloud computing, to derive protein structures with minimal effort. The portal eliminates the need to engage with intricate coding (like Python on a Jupyter notebook), enabling users to focus on protein inference results iterations.
The Google Cloud AlphaFold repository now includes the option to deploy this serverless portal, which offers a streamlined, secure, and centralized way to manage protein folding experiments. Launch new experiments with a single click, simplifying workflows and saving valuable time.
Centralized Pipelines
The portal makes researchers’ work more efficient in several ways:
- Centralized access: Multiple researchers can access the portal through a single web address instead of running their own Jupyter notebook instances or deploying infrastructure on separate projects.
- Streamlined protein folding: Researchers can run protein folding pipeline jobs under their usernames and filter simulation results based on other researchers’ work. This allows for easy comparison and fine-tuning.
- Enhanced collaboration: Previously, each researcher needed to run their own Jupyter notebook instance to run each protein-folding job. Now, they can collaborate more easily by accessing and comparing simulation results in a centralized location.

1- AlphaFold Portal Dashboard
Consider this dashboard to be the central hub for protein folding endeavors. Users can personalize the display, expertly filter results, and utilize designated link buttons to directly access protein resources. The need to navigate through complex configuration or executions has now been simplified.
Are you prepared to engage in protein folding? With just two clicks, your sequence (in FASTA format) will be processed and simulated. The UI will auto select recommendations for the optimal GPU machine configuration based on the type and size of your protein. However, if you are not satisfied with the suggested settings, you have the option to expand the advanced settings and customize them to your desired specifications.
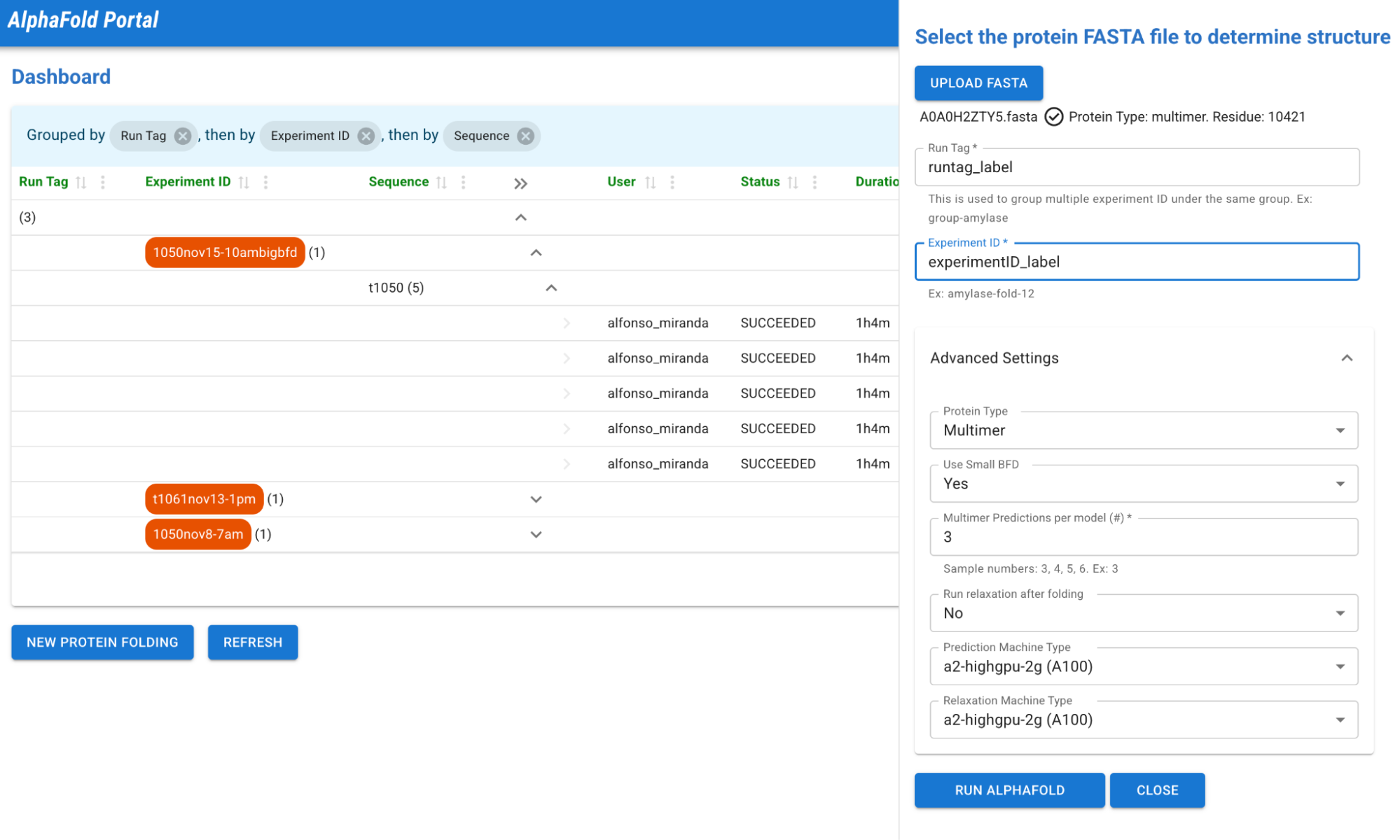
2 – New Protein Folding
Furthermore, we have integrated a preview function for your protein models. Tapping into an open-source visualization tool, you can now seamlessly explore the intricate molecular structures without leaving the interface.

3 – Protein structure visualization
This tool empowers everyone in your biotech organization to harness the power of protein folding, regardless of their cloud or coding experience. Executing this highly complex and compute intensive workload seamlessly on a streamlined, optimized infrastructure, ensuring efficiency and ease of use.
Getting started
If you’re a Google Cloud newbie, no worries! We recommend checking out the Getting Started page to get familiarized with Google Cloud. Then, create a project to house all this protein-folding magic.
To proceed, follow the instructions provided in the open-source Google Cloud AlphaFold repository, accessible via the link. This repository contains convenient, pre-built templates that will assist you in setting up all the necessary components. Kindly note that this part of the process may require some technical expertise. If you encounter any challenges or require guidance, your dedicated GCP representative is readily available to assist you in navigating the complexities of the cloud.