The FDA has a history of using real world evidence (RWE) as an integral component of the drug approval process. Moreover, RWE can mitigate the need for placebos in some clinical trials. The clinical records that make RWE evidence useful, however, often reside in unstructured formats, such as doctor’s notes, and must be “abstracted” into a clinical structured format. Cloud technologies and AI can help accelerate this process, making it significantly faster and more scalable.
Leading drug researchers are starting to augment their clinical trials with real world data for their FDA study submissions because it saves time and is more cost effective. Once the patient’s care concludes, the vast amounts of historical unstructured patient medical data ends up being a contributor to increasing storage needs. Unstructured data is key and critical in clinical decision support systems. In their original unstructured format, insights need a human to review the unstructured data. With no discrete data points from which insights can be quickly drawn, unstructured medical data can result in increased care gaps and care variances. Simple logic dictates that unassisted human abstraction alone is not fast or accurate enough to abstract all of this patient data. Applied natural language processing (NLP) using serverless software components on Google Cloud provides an efficient way of identifying and guiding clinical abstractors towards a prioritized list of patient medical documents.
How to run Medical Text Processing on Google Cloud
Using Google Cloud’s Vertex Workbench Jupyter Notebooks, you can create a data pipeline that takes raw clinical text documents and processes them through Google Cloud’s Healthcare Natural Language API landing the structured json output into BigQuery. From there, you can build a dashboard that can show clinical text characteristics, e.g., number of labels and relationships. From this, you’ll be able to build a trainable language model that can extract text and be further improved over time by human labeling.
To better understand how the solution addresses these challenges, let’s review the medical text entity extraction workflow:
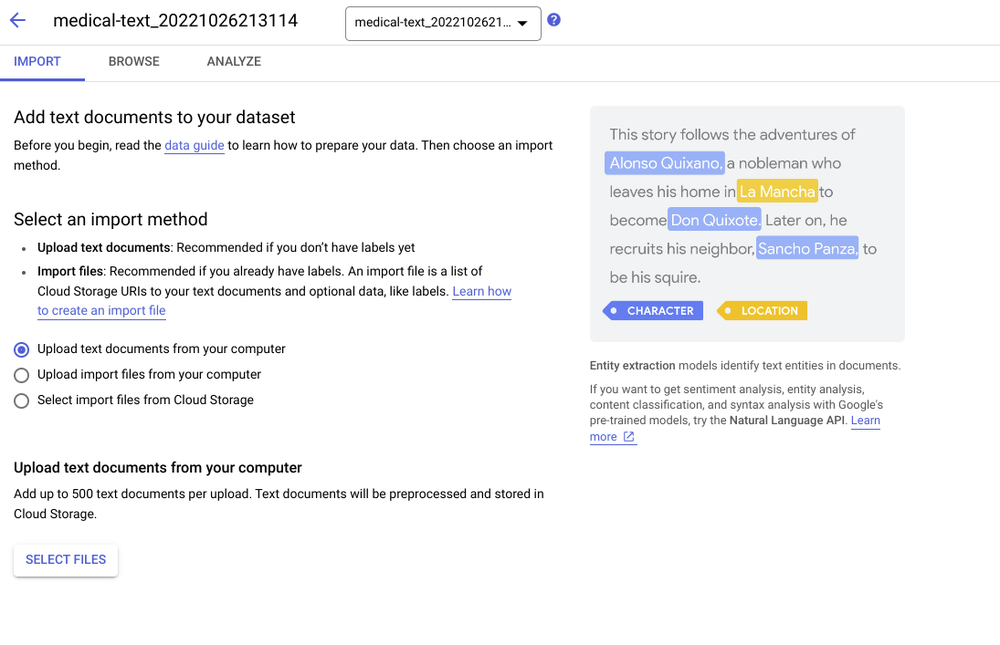
- Document AI for Data Ingestion. The system starts with a PDF file that contains de-identified medical text, such as a doctor’s hand-written notes or other unstructured text. This unstructured data is first processed by Document AI using optical character recognition (OCR) technology to digitize the text and images.
- Natural Language Processing. The Cloud Natural Language API includes a set of pretrained models, including models for extracting and classifying medical text. The labels that are generated as part of the output of this service will serve as the “ground truth” labels for the Vertex AI AutoML service where additional, domain specific custom labels will be added.
- Vertex AI AutoML. Vertex AI AutoML offers a machine learning toolset for human-in-the-loop dataset labeling and automatic label classification, using a Google model that your team can train with your data, even if team members possess little coding or data science expertise.
- BigQuery Tables. NLP processed records are stored in BigQuery for further processing and visualization.
- Looker Dashboard. The Looker Dashboard acts as the central “brain” for the clinical text abstraction process by serving visualizations that help the team identify the highest priority clinical documents using metrics like tag and concept “density.”
- Python Jupyter Notebook. Use either Colab (free) or Vertex AI (enterprise) notebooks to explore your text data and call different APIs for ingestion and NLP.
The Healthcare Natural Language API
The Healthcare Natural Language API lets you efficiently run medical text entity resolution at scale by focusing on the following optimizations:
- Optimizing document OCR and data extraction by using scalable Cloud Functions to run the document processing in parallel.
- Optimizing cost and time to market by using completely serverless and managed services.
- Facilitating a flexible and inclusive workflow that incorporates human-in-the-loop abstraction assisted by ML.
The following diagram shows the architecture of the solution.

- A set of reusable Python scripts that can be run from either a Jupyter notebook or Google Cloud Functions that drive the various stages of an NLP processing pipeline, which converts medical text to structured patient data and a Looker dashboard that acts as the decision support interface for teams of human clinical abstractors.
- A set of Google Cloud Storage Buckets to support the various stages of data processing (illustrated below).
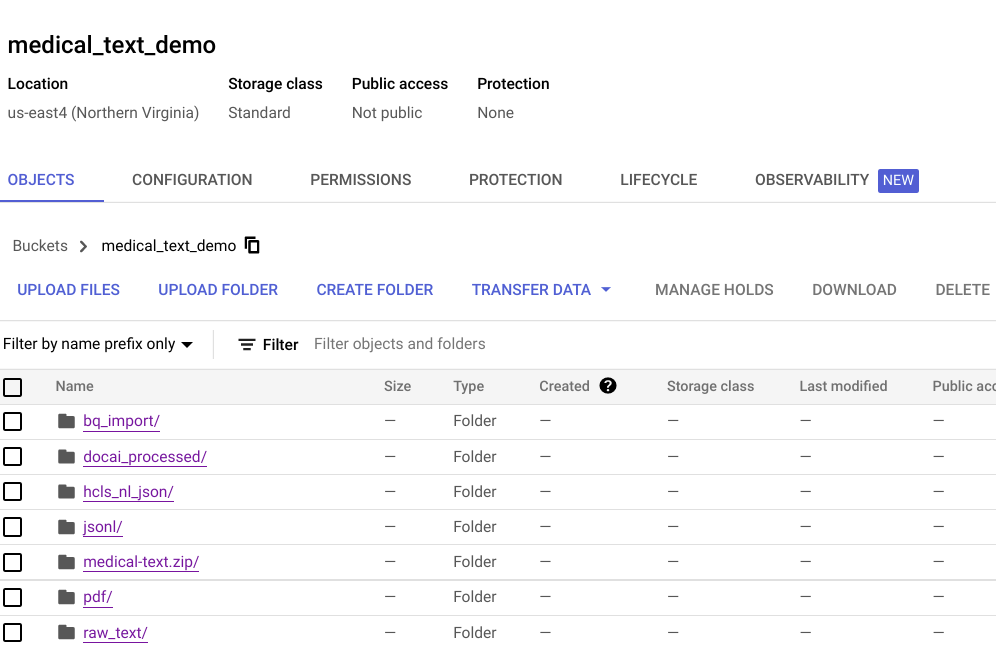
- Two BigQuery tables, called “Entity” and “Document,” in a dataset called “entity,” are created as the data model for the Looker dashboard.
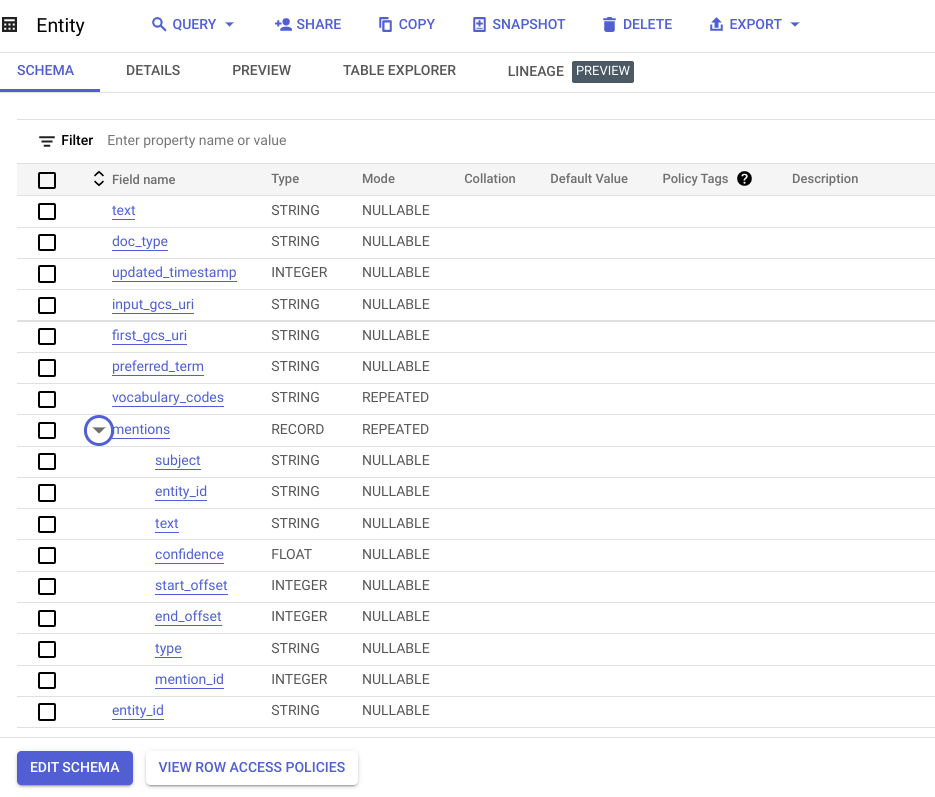
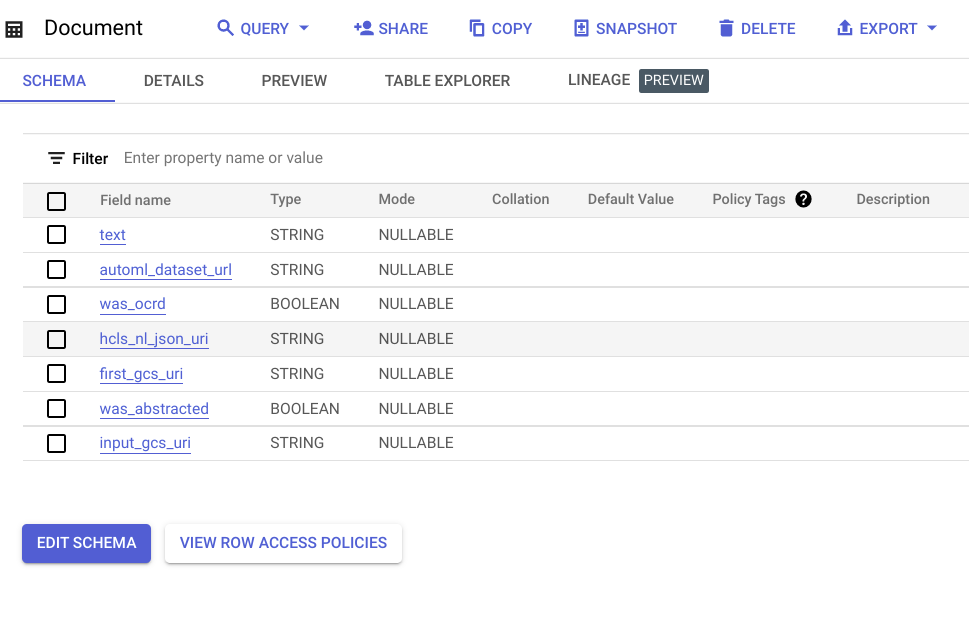
- A Vertex AI dataset used for human-in-the-loop labeling by clinical abstractors and to send labeling requests to the Google Vertex AI Labeling Team for added flexibility and scale.
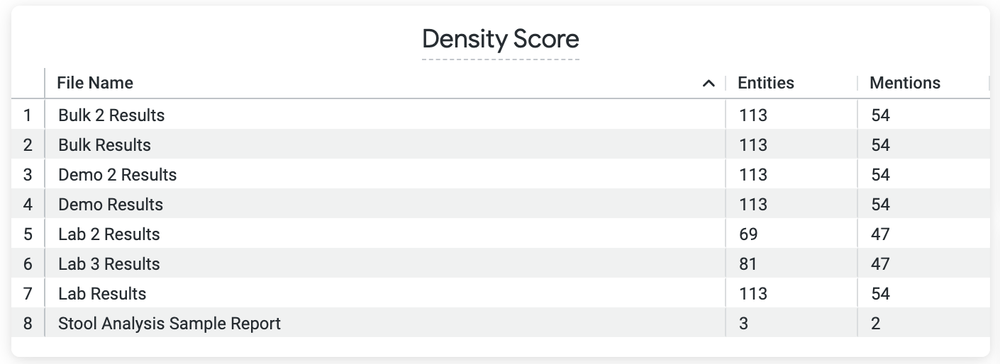
- A Looker dashboard that displays the stack-ranked documents to be processed in order by the human abstractors based on a custom “density” metric, which is the number of data elements (labels) found in those documents. This dashboard will guide the human abstractors to look at the sparsely labeled documents first and let Google’s NLP do the heavy lifting.
A list of documents, by density score, helps human abstractors know which documents need a lot of work versus only a light review.
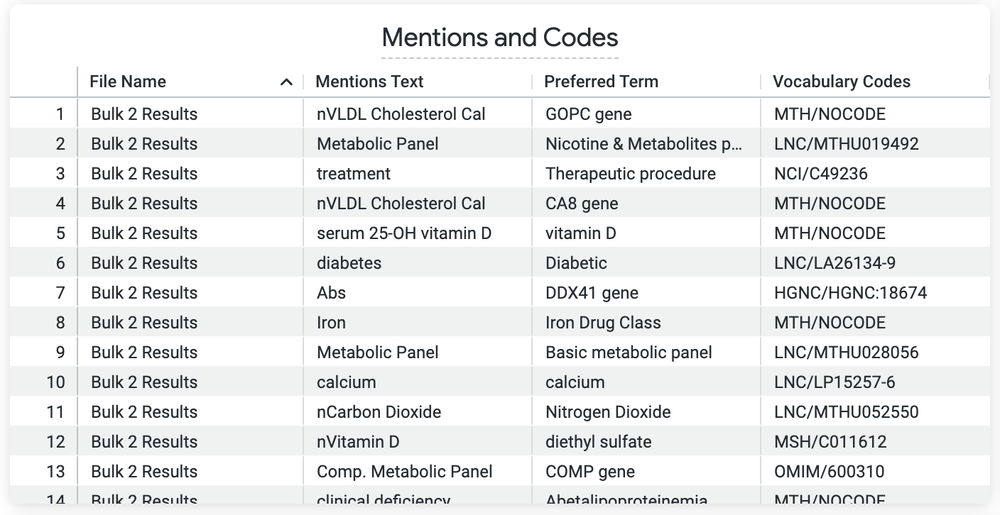
This Look (view) shows the coded medical text that was mapped to the UMLS clinical ontology by the Google Healthcare Natural Language API.
This Look (view) shows the entity mentions, including the subject of each mention and its confidence score, allowing for loading into a biomedical knowledge graph for further downstream analysis.

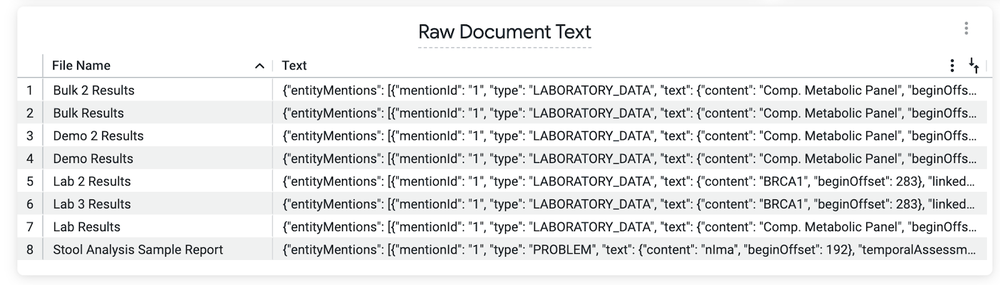
This Look (view) shows the entity mentions found in the raw document text.
Future Topics and Next Steps
This demo loaded the entity and document metadata into BigQuery and Looker but didn’t load the rich relationships that come out-of-the-box from the Healthcare Natural Language API. Using those relationships, it is possible to create a biomedical knowledge graph and explore the pathways between disease, treatment, and cohorts, and to help generate new hypotheses linking these facts.
We created a barebones dashboard with Looker. However, Looker has rich functionality, such as the ability to push to channels like chat when a document is available for review or to visualize the patient as a medical knowledge graph of related entities or embedding ML predictions right in the Looker LookML itself. This dashboard should be considered just a starting point for Looker powered clinical informatics.
To learn more about the Healthcare Natural Language API, please visit our product page. To try it yourself for free, please visit this demo link.
For help with loading this example medical text into a Vertex AI dataset for labeling, please contact the Google Cloud Biotech Team.
Data Privacy
No real patient data was used for any part of this blog post. Google Cloud’s customers retain control over their data. In healthcare settings, access and use of patient data is protected through the implementation of Google Cloud’s reliable infrastructure and secure data storage that support HIPAA compliance, along with each customer’s security, privacy controls, and processes. To learn more about data privacy on Google Cloud, check out this link.